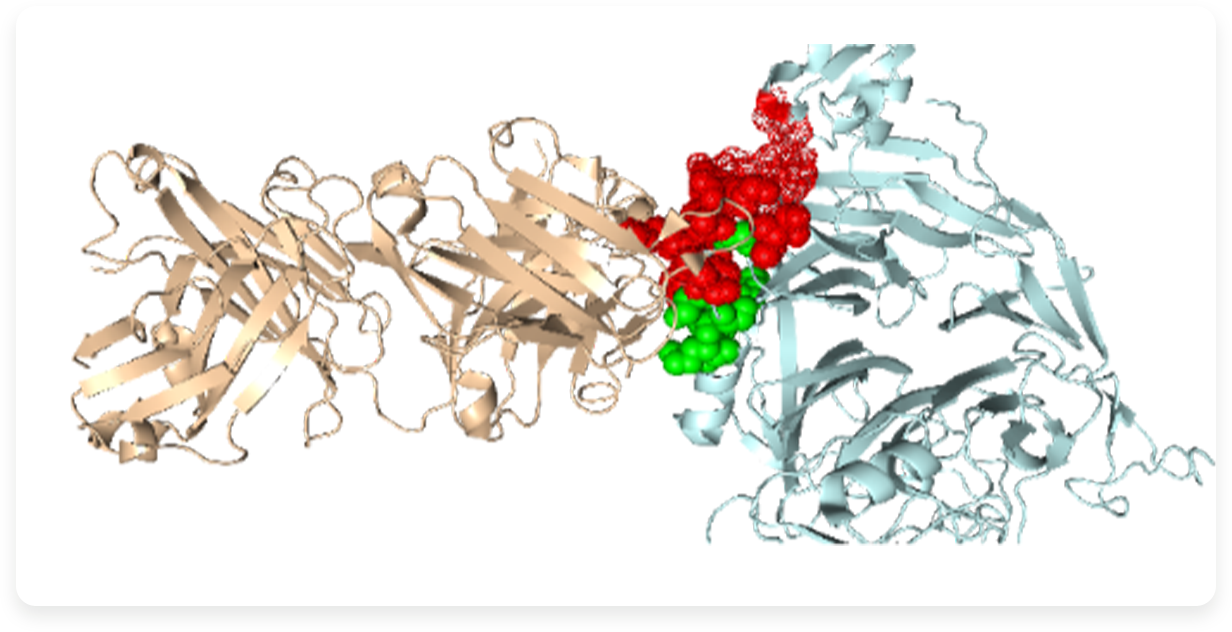
Antibody recognition epitopes (Epitopes) are the core characterization of antibodies, an important dimension for antibody evaluation, and a key element of antibody function. AbMapTM high-throughput antibody recognition epitope analysis technology, combined with a universal phage display peptide library, high-throughput sequencing, and AI algorithm, can quickly, accurately, low-cost, and high-throughput identify antibody recognition epitopes and provide core amino acid information.
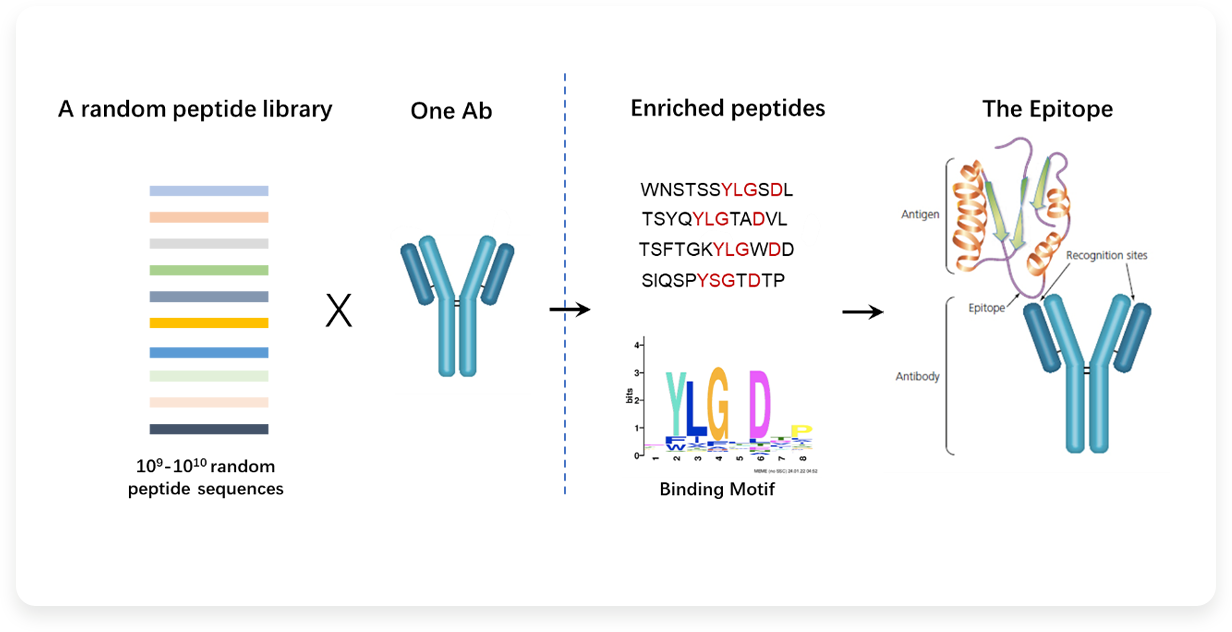
Based on a universal, highly diverse, 12aa long phage random peptide library, AbMap can quickly detect peptide segments recognized by antibodies and obtain their amino acid sequence patterns or shared motifs through algorithms. The identified epitopes can be obtained by primary Sequence alignment or three-dimensional conformational reconstruction. At the same time, the common motif can accurately extract key amino acids and the preference of antibody Recognition sequence. NGS can achieve parallel detection and analysis of multiple antibodies.
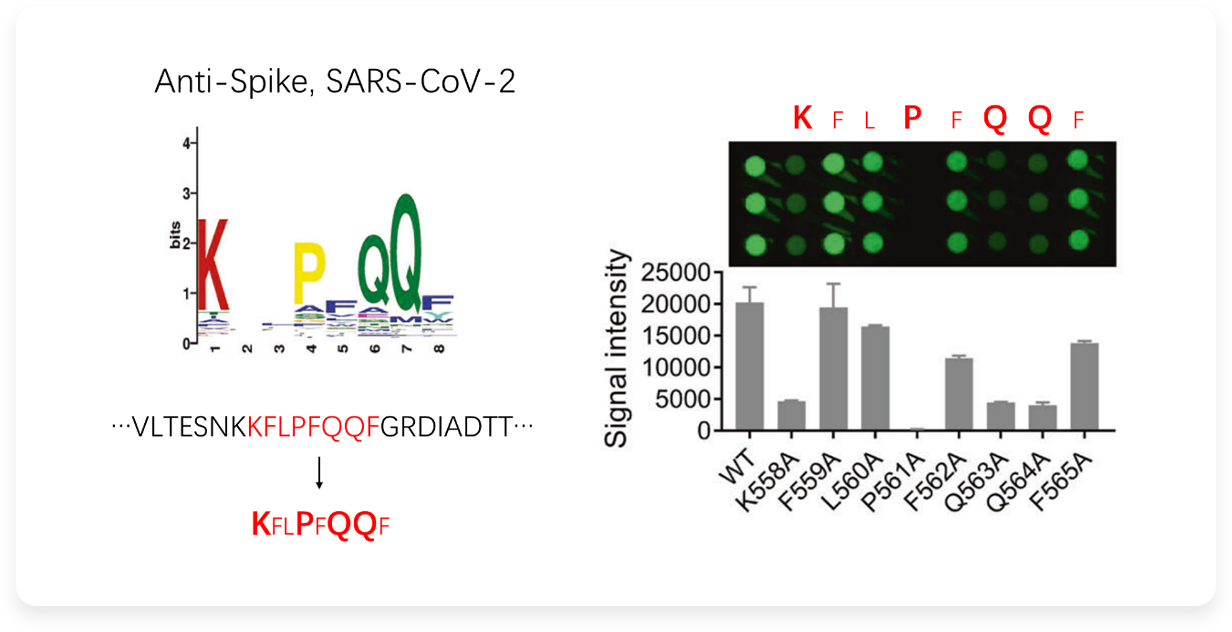
For an antibody against the spike of COVID-19, the recognized amino acid motif was first obtained through AbMap, and its linear epitope (red labeled sequence) was obtained through Sequence alignment. At the same time, it was suggested that K, P, Q and Q were core amino acids. In order to verify this result, several polypeptides with single Point mutation of amino acids were synthesized, and a polypeptide chip was constructed to detect this antibody. The results were consistent with expectations. Mutations of key amino acids greatly weakened the binding of antibodies, while mutations of other amino acids had little impact.
Monoclonal antibody, polyclonal antibody;
Full length IgG, Fab;
Human, mouse, rabbit, etc


Antibody concentration:>0.5 mg/mL;
Purity:>85%;
Total:>1 μ G